Abstract
Cerebral cavernous malformations (CCMs) are common inherited and sporadic vascular malformations that cause strokes and seizures in younger individuals1. CCMs arise from endothelial cell loss of KRIT1, CCM2 or PDCD10, non-homologous proteins that form an adaptor complex2. How disruption of the CCM complex results in disease remains controversial, with numerous signalling pathways (including Rho3,4, SMAD5 and Wnt/β-catenin6) and processes such as endothelial–mesenchymal transition (EndMT)5 proposed to have causal roles. CCM2 binds to MEKK3 (refs 7, 8, 9, 10, 11), and we have recently shown that CCM complex regulation of MEKK3 is essential during vertebrate heart development12. Here we investigate this mechanism in CCM disease pathogenesis. Using a neonatal mouse model of CCM disease, we show that expression of the MEKK3 target genes Klf2 and Klf4, as well as Rho and ADAMTS protease activity, are increased in the endothelial cells of early CCM lesions. By contrast, we find no evidence of EndMT or increased SMAD or Wnt signalling during early CCM formation. Endothelial-specific loss of Map3k3 (also known as Mekk3), Klf2 or Klf4 markedly prevents lesion formation, reverses the increase in Rho activity, and rescues lethality. Consistent with these findings in mice, we show that endothelial expression of KLF2 and KLF4 is increased in human familial and sporadic CCM lesions, and that a disease-causing human CCM2 mutation abrogates the MEKK3 interaction without affecting CCM complex formation. These studies identify gain of MEKK3 signalling and KLF2/4 function as causal mechanisms for CCM pathogenesis that may be targeted to develop new CCM therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cavalcanti, D. D. et al. Cerebral cavernous malformations: from genes to proteins to disease. J. Neurosurg. 116, 122–132 (2012)
Plummer, N. W., Zawistowski, J. S. & Marchuk, D. A. Genetics of cerebral cavernous malformations. Curr. Neurol. Neurosci. Rep. 5, 391–396 (2005)
Whitehead, K. J. et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nature Med. 15, 177–184 (2009)
Stockton, R. A., Shenkar, R., Awad, I. A. & Ginsberg, M. H. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 207, 881–896 (2010)
Maddaluno, L. et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature 498, 492–496 (2013)
Bravi, L. et al. Sulindac metabolites decrease cerebrovascular malformations in CCM3-knockout mice. Proc. Natl Acad. Sci. USA 112, 8421–8426 (2015)
Uhlik, M. T. et al. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nature Cell Biol. 5, 1104–1110 (2003)
Zawistowski, J. S. et al. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum. Mol. Genet. 14, 2521–2531 (2005)
Cullere, X., Plovie, E., Bennett, P. M., MacRae, C. A. & Mayadas, T. N. The cerebral cavernous malformation proteins CCM2L and CCM2 prevent the activation of the MAP kinase MEKK3. Proc. Natl Acad. Sci. USA (2015)
Fisher, O. S. et al. Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex. Nature Commun. 6, 7937 (2015)
Wang, X. et al. Structural insights into the molecular recognition between cerebral cavernous malformation 2 and mitogen-activated protein kinase kinase kinase 3. Structure 6, 1087–1096 (2015)
Zhou, Z. et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev. Cell 32, 168–180 (2015)
Boulday, G. et al. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J. Exp. Med. 208, 1835–1847 (2011)
Glading, A., Han, J., Stockton, R. A. & Ginsberg, M. H. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J. Cell Biol. 179, 247–254 (2007)
Wüstehube, J. et al. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc. Natl Acad. Sci. USA 107, 12640–12645 (2010)
Cuttano, R. et al. KLF4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol. Med. 8, 6–24 (2015)
Ferrer-Vaquer, A. et al. A sensitive and bright single-cell resolution live imaging reporter of Wnt/β-catenin signaling in the mouse. BMC Dev. Biol. 10, 121 (2010)
Whitehead, K. J., Plummer, N. W., Adams, J. A., Marchuk, D. A. & Li, D. Y. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development 131, 1437–1448 (2004)
Gibson, C. C. et al. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation 131, 289–299 (2015)
Fisher, O. S. et al. Structural basis for the disruption of the cerebral cavernous malformations 2 (CCM2) interaction with Krev interaction trapped 1 (KRIT1) by disease-associated mutations. J. Biol. Chem. 290, 2842–2853 (2015)
Draheim, K. M. et al. CCM2–CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J. Cell Biol. 208, 987–1001 (2015)
Spiegler, S. et al. High mutation detection rates in cerebral cavernous malformation upon stringent inclusion criteria: one-third of probands are minors. Mol. Genet. Genomic Med. 2, 176–185 (2014)
Marchi, S. et al. Defective autophagy is a key feature of cerebral cavernous malformations. EMBO Mol. Med. 7, 1403–1417 (2015)
Borikova, A. L. et al. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J. Biol. Chem. 285, 11760–11764 (2010)
Zheng, X. et al. Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev. Cell 23, 342–355 (2012)
Pagenstecher, A., Stahl, S., Sure, U. & Felbor, U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum. Mol. Genet. 18, 911–918 (2009)
Akers, A. L., Johnson, E., Steinberg, G. K., Zabramski, J. M. & Marchuk, D. A. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 18, 919–930 (2009)
Chao, T. H., Hayashi, M., Tapping, R. I., Kato, Y. & Lee, J. D. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J. Biol. Chem. 274, 36035–36038 (1999)
McDonald, D. A. et al. Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke 43, 571–574 (2012)
Wang, Y. et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486 (2010)
Mleynek, T. M. et al. Lack of CCM1 induces hypersprouting and impairs response to flow. Hum. Mol. Genet. 23, 6223–6234 (2014)
Lee, J. S. et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev. Cell 11, 845–857 (2006)
Katz, J. P. et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129, 2619–2628 (2002)
Kisanuki, Y. Y. et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230, 230–242 (2001)
Acknowledgements
We thank the members of the Kahn laboratory for their comments during the course of this work. These studies were supported by National Institutes of Health (NIH) grants R01HL094326 (to M.L.K.), P01NS092521 (to I.A.A. and M.L.K.), VAMC 1IO1BX002976 (to D.Y.L.), RO1HL-084516 (to D.Y.L.), R01NS075168 (to K.J.W.), T32HL07439 (to A.T.T.), and Australian NHMRC project grant 161558 (to X.Z.).
Author information
Authors and Affiliations
Contributions
Z.Z., A.T.T. and W.-Y.W. designed and performed most of the experiments and helped to write the manuscript. S.B. performed all of the microCT CCM lesion measurements in a blinded manner. J.S.C.A., K.J.W., R.S., I.A.A. and D.Y.L. provided critical reagents. S.B., L.M.G., S.Z., J.Y., A.C.W., M.F., X.Z. and M.L.K. helped to design and perform the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
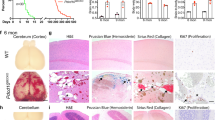
Extended Data Figure 1 Histological characteristics of early CCM lesions and the cerebellar white matter in which they form.
a, P7 and P8 CCM lesions in the Krit1 model. Hindbrains from Krit1ECKO mice were collected at P7 and P8, and sections were stained with haematoxylin and eosin to detect vascular malformations. Images are representative of 4 independent experiments. Dotted lines indicate the cerebellar white matter; arrows indicate CCM lesions. b, Versican is abundant in the white matter of the P7 hindbrain, the site of primary CCM formation in the neonatal mouse model. Haematoxylin and eosin staining of the hindbrain at low (top left) and high (bottom left) magnifications. Antibody staining reveals abundant versican protein in the white matter of the hindbrain (centre), the site at which CCM lesions specifically appear at this time point in the Krit1 mouse model, and less abundant versican in the cortex. DPEAAE staining reveals a pattern of ADAMTS-mediated versican degradation that is the inverse of intact versican; that is, higher in the cortex and lower in the white matter (right). Results are representative of 3 independent experiments. Scale bars, 500 μm (top) and 100 μm (bottom). Boxes indicate regions shown at higher magnification below; dotted lines indicate the cerebellar white matter.
Extended Data Figure 2 Endothelial Rho activity, but not β-catenin or SMAD3 signalling, is increased during CCM formation.
a, Immunostaining for the endothelial marker PECAM and pMLC in the white matter of the cerebellum of P6 control and Krit1ECKO littermates. Scale bars, 50 μm. b, Anti-GFP immunostaining to detect TCF/Lef:H2B-GFP Wnt/β-catenin reporter (WntREP) activity. Scale bars, 50 μm. c, Immunoblotting of P6 brain endothelial cell lysate for GFP. Results are representative of 3 independent experiments. d, qPCR analysis of β-catenin target genes in hindbrain endothelial cells (n = 4–5). P > 0.05 (unpaired two-tailed Student’s t-test) for comparison of all values. Error bars indicate s.e.m. e, Immunostaining for phosphorylated-SMAD3 (pSMAD3) and PECAM. Scale bars, 50 μm. f, Total SMAD3 and pSMAD3 were detected by immunoblotting cerebellar endothelial cell lysates. Results are representative of 3 independent experiments.
Extended Data Figure 3 Hes1 expression rises after primary CCM lesion formation in Krit1ECKO hindbrain endothelial cells, and is reversed by Map3k3 haploinsufficiency.
a, qPCR analysis of Notch target genes in endothelial cells freshly collected from the hindbrain of P6 or P11 Krit1ECKO animals compared with control, tamoxifen-treated littermates (n = 4). **P < 0.01 (unpaired two-tailed Student’s t-test). b, qPCR analysis of gene expression in endothelial cells freshly collected from the cerebellum of P11 Krit1fl/fl and Krit1ECKO animals, with and without endothelial loss of one Map3k3 allele (n = 4). **P < 0.01 (unpaired two-tailed Student’s t-test). Error bars indicate s.e.m.
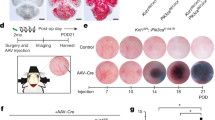
Extended Data Figure 4 Map3k3 haploinsufficiency rescues both early embryonic lethality due to endothelial loss of Krit1 and postnatal CCM lesion formation due to endothelial loss of Ccm2.
a, Partial loss of MEKK3 rescues early lethality associated with endothelial deletion of Krit1. E10.5 littermate embryos are shown at the same magnification. Tie2Cre;Krit1fl/fl animals lacking endothelial KRIT1 die by E9.5 due to lack of patent branchial arch arteries (middle). By contrast, Tie2Cre;Krit1fl/fl;Map3k3fl/+ animals develop patent arteries and survive past mid-gestation (right). Note the presence of pericardial oedema due to a persistent cardiac defect in the Tie2Cre;Krit1fl/fl;Map3k3fl/+ embryo. Images are representative of 3 independent experiments. b, Rescue of CCM formation in the Ccm2 model with loss of MEKK3. iECre;Ccm2fl/fl;Map3k3+/+ (Ccm2ECKO) and iECre;Ccm2fl/fl;Map3k3fl/+ neonates were induced with tamoxifen at P1 and lesion formation scored by visual detection in the hindbrain at P11 as done for Krit1ECKO animals. Images are representative of 4 independent experiments. Scale bars, 1 mm.
Extended Data Figure 5 Rescue of CCM formation in Krit1ECKO animals with homozygous loss of Klf2.
Haematoxylin and eosin sections through the hindbrain of P11 iECre;Krit1fl/fl;Klf2fl/fl and control iECre;Krit1fl/fl animals are shown. No true CCM lesions were observed but venous dilatation was evident (arrow). Images are representative of 3 independent experiments. Dotted lines indicate the cerebellar white matter. Scale bars, 500 μm (left), 100 μm (middle) and 50 μm (right).
Extended Data Figure 6 CCM2 binds MEKK3 via its C-terminal helical harmonin domain.
a, MEKK3 binds the C-terminal domain of CCM2. Top, schematic of the CCM2 protein domains. Bottom, series of N-terminal-truncated CCM2–HA proteins used to map the CCM2–MEKK3 binding region. MEKK3–Flag and the indicated N-terminally truncated CCM2–HA proteins were co-expressed in HEK293T cells before immunoprecipitation with anti-HA antibodies and immunoblotting with anti-Flag antibodies to detect associated MEKK3 proteins (top). Immunoprecipitated HA–CCM2 was detected with anti-HA antibodies (below). b, MEKK3–Flag and the indicated C-terminally truncated CCM2–HA proteins were co-immunoprecipitated to map the CCM2–MEKK3 binding region. c, CCM2 CCCTdup protein is stably expressed in cultured endothelial cells. CCM2 CCCTdup–HA protein was expressed in cultured HUVECs using a tetracycline-inducible lentiviral vector at varying doses of doxycycline. The total (that is, endogenous CCM2 plus viral CCM2 CCCTdup) CCM2 mRNA levels were measured using qPCR (bottom), and CCM2 CCCTdup protein detected using anti-HA immunoblotting (top). GAPDH immunoblots are shown for loading control.
Extended Data Figure 7 The versican-degrading ADAMTS proteases are regulated by KLF2 and KLF4.
a, siRNA knockdown of KLF2 or KLF4 in cultured HUVECs reduces ADAMTS4 expression, as measured by qPCR. b, Adenoviral expression of KLF2 drives expression of the versican-degrading proteases ADAMTS1, ADAMTS4 and ADAMTS9 in addition to the known KLF2/4 target gene eNOS (also known as NOS3). KLF4 levels are reduced in KLF2-overexpressing HUVECs. Results are shown relative to expression following exposure to control adeno-LacZ virus. c, Adenoviral expression of KLF4 drives expression of ADAMTS4 in addition to eNOS. KLF2 levels are reduced in KLF4-overexpressing HUVECs. Results are shown relative to expression after exposure to control adeno-LacZ virus (n = 4–5). *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired two-tailed Student’s t-test). Error bars indicate s.e.m.
Extended Data Figure 8 The MEK5 inhibitor BIX02189 and the ERK5 inhibitor XMD17-109 reverse the increase in KLF2 and KLF4 expression conferred by loss of KRIT1 in cultured endothelial cells.
HUVECs were treated with scrambled or KRIT1-targeted siRNAs with or without BIX02189 (10 μM) and XMD17-109 (1 μM) (MEK5 and ERK5 inhibitors, respectively) for 24 h, and the levels of KLF2 and KLF4 mRNA were measured by qPCR (n = 3). **P < 0.01; ***P < 0.001 (unpaired two-tailed Student’s t-test). Error bars indicate s.e.m.
Extended Data Figure 9 Imaging and volume measurement of mouse hindbrain CCM lesions using microCT.
The imaging of P11 mouse hindbrains and analysis of the raw data to create composite CCM lesion images and volumes are shown.
Supplementary information
Supplementary Figure 1
This file contains uncropped western blot scans with size indications corresponding to Figures 1f, 2c, 2f, 5d, 5e and Extended Data Figures 5a, 5b, 5c. (PDF 4725 kb)
Rights and permissions
About this article
Cite this article
Zhou, Z., Tang, A., Wong, WY. et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3–KLF2/4 signalling. Nature 532, 122–126 (2016). https://doi.org/10.1038/nature17178
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17178
This article is cited by
-
Single-nucleus DNA sequencing reveals hidden somatic loss-of-heterozygosity in Cerebral Cavernous Malformations
Nature Communications (2023)
-
Single-cell sequencing reveals that endothelial cells, EndMT cells and mural cells contribute to the pathogenesis of cavernous malformations
Experimental & Molecular Medicine (2023)
-
Impaired retinoic acid signaling in cerebral cavernous malformations
Scientific Reports (2023)
-
Genetics of brain arteriovenous malformations and cerebral cavernous malformations
Journal of Human Genetics (2023)
-
A novel KRIT1/CCM1 mutation accompanied by a NOTCH3 mutation in a Chinese family with multiple cerebral cavernous malformations
neurogenetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.