Abstract
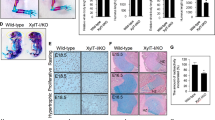
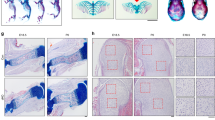
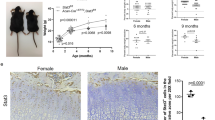
Perlecan, a large, multi-domain, heparan sulfate proteoglycan originally identified in basement membrane, interacts with extracellular matrix proteins, growth factors and receptors, and influences cellular signalling1,2,3,4,5. Perlecan is present in a variety of basement membranes and in other extracellular matrix structures5,6. We have disrupted the gene encoding perlecan (Hspg2) in mice. Approximately 40% of Hspg2–/– mice died at embryonic day (E) 10.5 with defective cephalic development. The remaining Hspg2–/– mice died just after birth with skeletal dysplasia characterized by micromelia with broad and bowed long bones, narrow thorax and craniofacial abnormalities. Only 6% of Hspg2–/– mice developed both exencephaly and chondrodysplasia. Hspg2–/– cartilage showed severe disorganization of the columnar structures of chondrocytes and defective endochondral ossification. Hspg2–/– cartilage matrix contained reduced and disorganized collagen fibrils and glycosaminoglycans, suggesting that perlecan has an important role in matrix structure. In Hspg2–/– cartilage, proliferation of chondrocytes was reduced and the prehypertrophic zone was diminished. The abnormal phenotypes of the Hspg2–/– skeleton are similar to those of thanatophoric dysplasia (TD) type I, which is caused by activating mutations in FGFR3 (refs 7, 8, 9), and to those of Fgfr3 gain-of-function mice10,11. Our findings suggest that these molecules affect similar signalling pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Noonan, D.M. et al. The complete sequence of perlecan, a basement membrane heparan sulfate proteoglycan, reveals extensive similarity with laminin A chain, low density lipoprotein-receptor, and the neural cell adhesion molecule. J. Biol. Chem. 266, 22939–22947 (1991).
Iozzo, R.V., Cohen, I.R., Grassel, S. & Murdoch, A.D. The biology of perlecan: the multifaceted heparan sulphate proteoglycan of basement membranes and pericellular matrices. Biochem. J. 302, 625–639 (1994).
Aviezer, D. et al. Perlecan, basal lamina proteoglycan, promotes basic fibroblast growth factor-receptor binding, mitogenesis, and angiogenesis. Cell 79, 1005–1013 ( 1994).
SundarRaj, N., Fite, D., Ledbetter, S., Chakravarti, S. & Hassell, J.R. Perlecan is a component of cartilage matrix and promotes chondrocyte attachment. J. Cell. Sci. 108, 2663–2672 (1995).
Handler, M., Yurchenco, P.D. & Iozzo, R.V. Developmental expression of perlecan during murine embryogenesis. Dev. Dyn. 210, 130– 145 (1997).
Morriss-Kay, G. & Tucket, F. Early events in mammalian craniofacial morphogenesis. J. Craniofac. Genet. Dev. Biol. 11, 181–191 ( 1991).
Tavormina, P.L. et al. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nature Genet. 9, 321–328 ( 1995).
Horton, W.A. Fibroblast growth factor receptor 3 and the human chondrodysplasias. Curr. Opin. Pediatr. 9, 437–442 (1997).
Weber, M., Johannisson, T., Thomsen, M., Rehder, H. & Niethard, F.U. Thanatophoric dysplasia type I: new radiologic, morphologic, and histologic aspects toward the exact definition of the disorder. J. Pediatr. Orthop. B 7, 1–9 (1998).
Naski, M.C., Colvin, J.S., Coffin, J.D. & Ornitz, D.M. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development 125, 4977–4988 (1998).
Li, C. et al. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum. Mol. Genet. 8, 35– 44 (1999).
Wilcox, W.R. et al. Molecular, radiologic, and histopathologic correlations in thanatophoric dysplasia. Am. J. Med. Genet. 78, 274–281 (1998).
Naski, M.C., Wang, Q., Xu, J. & Ornitz, D.M. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nature Genet. 13, 233–237 (1996).
Chuang, P.T. & McMahon, A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397, 617–621 (1999).
Aszodi, A., Pfeifer, A., Wendel, M., Hiripi, L. & Fassler, R. Mouse models for extracellular matrix diseases. J. Mol. Med. 76, 238–252 (1998).
Watanabe, H. & Yamada, Y. Mice lacking link protein develop dwarfism and craniofacial abnormalities. Nature Genet. 21, 225–229 (1999).
Deng, C., Wynshaw-Boris, A., Zhou, F., Kuo, A. & Leder, P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84, 911–921 (1996).
Ornoy, A., Adomian, G.E., Eteson, D.J., Burgeson, R.E. & Rimoin, D.L. The role of mesenchyme-like tissue in the pathogenesis of thanatophoric dysplasia. Am. J. Med. Genet. 21, 613–630 ( 1985).
Horton, W.A., Hood, O.J., Machado, M.A., Ahmed, S. & Griffey, E.S. Abnormal ossification in thanatophoric dysplasia. Bone 9, 53–61 (1988).
Vortkamp, A. et al. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273, 613–622 (1996).
Colvin, J.S., Bohne, B.A., Harding, G.W., McEwen, D.G. & Ornitz, D.M. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nature Genet. 12, 390–397 ( 1996).
Su, W.C. et al. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature 386, 288–292 (1997).
Sahni, M. et al. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes. Dev. 13, 1361–1366 (1999).
Ornitz, D.M. et al. FGF binding and FGF receptor activation by synthetic heparan-derived di- and trisaccharides. Science 268, 432 –436 (1995).
Delezoide, A.L. et al. Abnormal FGFR3 expression in cartilage of thanatophoric dysplasia fetuses. Hum. Mol. Genet. 6, 1899 –1906 (1997).
Aviezer, D., Iozzo, R.V., Noonan, D.M. & Yayon, A. Suppression of autocrine and paracrine functions of basic fibroblast growth factor by stable expression of perlecan antisense cDNA. Mol. Cell. Biol. 17, 1938–1946 ( 1997).
Tybulewicz, V.L., Crawford, C.E., Jackson, P.K., Bronson, R.T. & Mulligan, R.C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65, 1153–1163 ( 1991).
Peters, P.W. Double staining of fetal skeletons for cartilage and bone. in Methods in Prenatal Toxicology(eds Neubert, D. et al.) 153–154 (Thieme, Stuttgart, 1977).
Arahata, K. et al. Immunostaining of skeletal and cardiac muscle surface membrane with antibody against Duchenne muscular dystrophy peptide. Nature 333, 861–863 ( 1988).
Harris, M.J. & Juriloff, D.M. Genetic landmarks for defects in mouse neural tube closure. Teratology 56, 177–187 (1997).
Acknowledgements
We thank G. Longenecker for blastocyst injection; W. Swaim, N. Marinos, V. Morgan, L. Bowers and R. Yaskovich for histology and electron microscopy; M. Mankani for Faxitron radiographs; J. Sasse for Fgf1 antibody; C. Deng for Fgfr3 cDNA; A. McMahon for Ihh cDNA; G. Lunstrum and N. Morris for collagen X antibody; A. Kulkarni, S. Kimura, T. Oshima, M. Hirasawa and K. Kimata for technical comments; and H. Kleinman for critically reading the manuscript. Some of this work was supported by a grant from Seikagaku Corporation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arikawa-Hirasawa, E., Watanabe, H., Takami, H. et al. Perlecan is essential for cartilage and cephalic development. Nat Genet 23, 354–358 (1999). https://doi.org/10.1038/15537
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/15537
This article is cited by
-
Basal lamina changes in neurodegenerative disorders
Molecular Neurodegeneration (2021)
-
Knockdown of hspg2 is associated with abnormal mandibular joint formation and neural crest cell dysfunction in zebrafish
BMC Developmental Biology (2021)
-
Association between rare variants in specific functional pathways and human neural tube defects multiple subphenotypes
Neural Development (2020)
-
Assembling custom side chains on proteoglycans to interrogate their function in living cells
Nature Communications (2020)
-
Versican is crucial for the initiation of cardiovascular lumen development in medaka (Oryzias latipes)
Scientific Reports (2019)